Principle of CETSA
CETSA®
CETSA builds on the fundamental principle that a protein’s thermal stability changes when its environment is altered—such as when a small molecule, drug, or ligand binds to its target. This interaction increases or decreases the protein’s heat resistance, creating a measurable thermal shift in its melt profile.
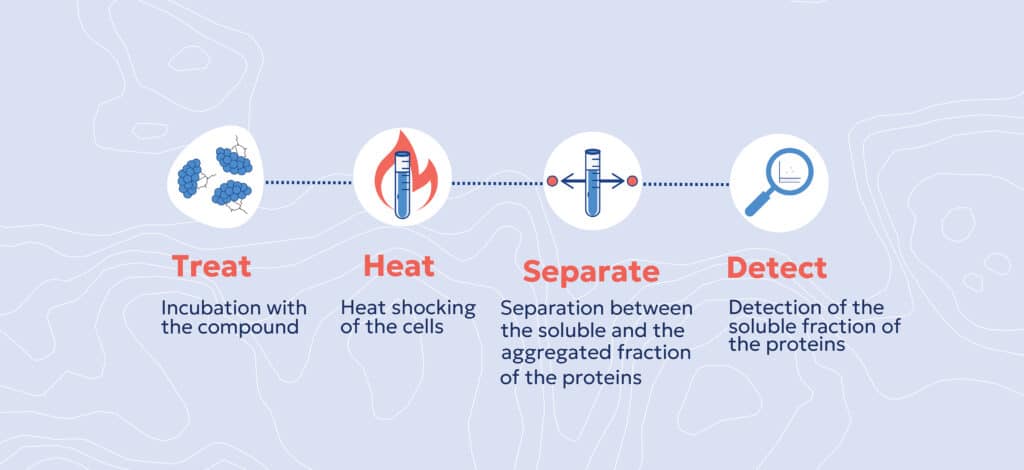
How CETSA Works
- Incubation – Cells, tissues, or lysates are treated with or without the test compound.
- Heat Shock – A controlled increase in temperature induces protein denaturation.
- Thermal Stability Measurement – The soluble (non-denatured) protein fraction is quantified across different temperature conditions.
- Target Engagement Confirmation – If a compound binds to its target, it alters the protein’s heat resistance, producing a detectable thermal shift.
By measuring these direct binding events in a native biological environment, CETSA provides a high-confidence assessment of target engagement, independent of enzymatic activity or artificial assay conditions.
Advantages of CETSA Over Traditional Assays
- Native Cellular Context – CETSA directly measures protein stability and drug-target interactions in intact cells, offering more biologically relevant data than in vitro biochemical assays.
- Label-Free Approach – No need for chemical labeling or reporter tags, preserving natural protein behavior and reducing assay artifacts.
- No Genetic Modification Required – CETSA does not require tagged proteins or genetically modified cell lines unlike some target engagement assays.
- Broad Applicability – CETSA can be applied to virtually any protein, regardless of function, enzymatic activity, or availability of specific antibodies.
- Direct Measurement of Drug Binding – Rather than relying on indirect readouts like enzymatic activity or reporter gene expression, CETSA quantifies direct stabilization or destabilization of proteins upon ligand binding.
CETSA has become an indispensable tool in modern drug discovery research by enabling high-confidence, label-free drug-target interaction studies.
Explore our CETSA Application Notes

Is Your Hit on Target?
Validate Early with CETSA® Accelerate hit confirmation and lead optimization with CETSA® …

The Power of Proteome-wide CETSA®
Uncover Drug Mechanisms with Confidence Unlock deeper insights into drug mechanisms with …
Is Your Compound Actually Therapeutic?
Translatable Insights with CETSA® Translatable Insights with CETSA® on CDK4 and RIPK…

