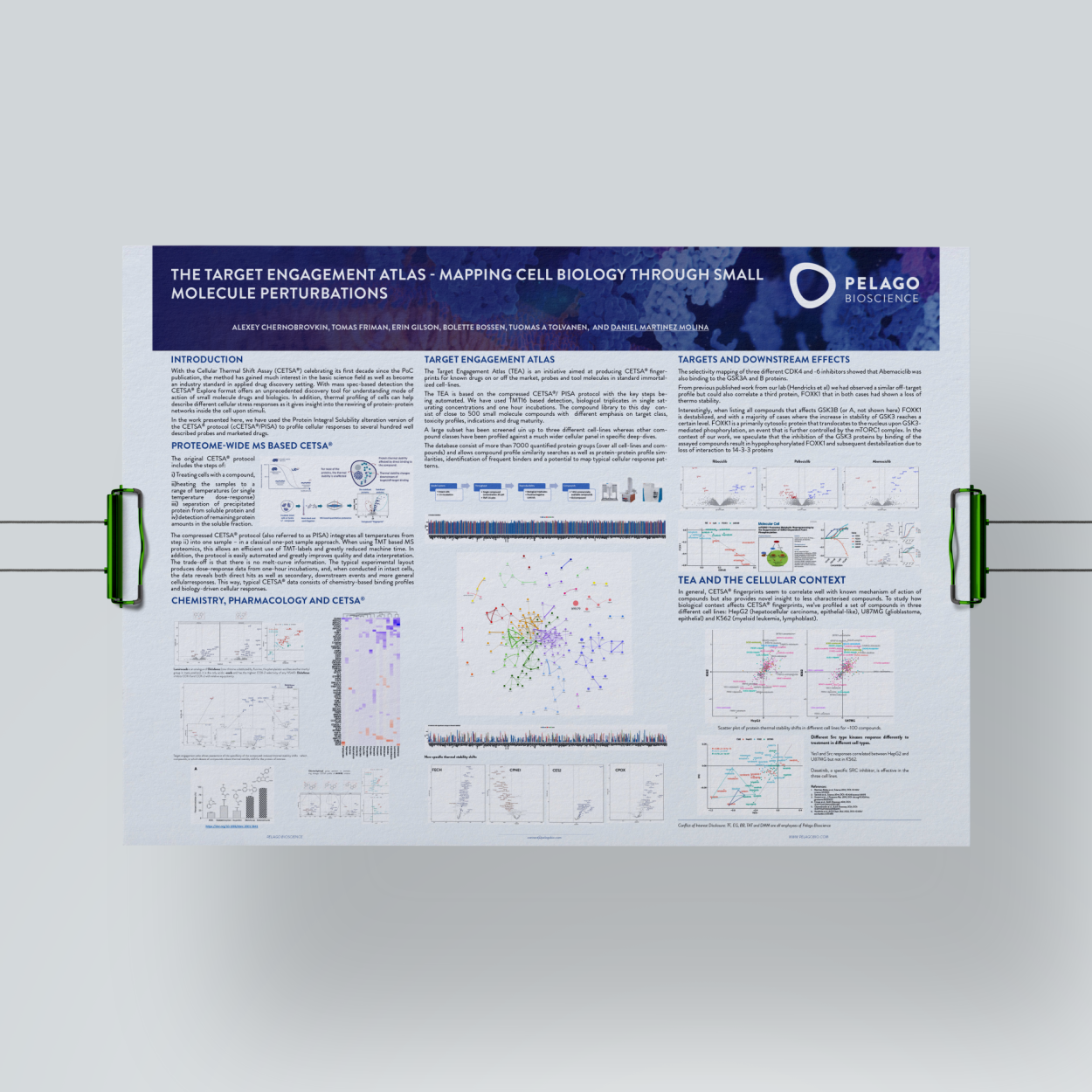
The Target Engagement Atlas – Mapping Cell Biology Through Small Molecule Perturbations
INTRODUCTION
With the Cellular Thermal Shift Assay (CETSA®) celebrating its first decade since the PoC publication, the method has gained much interest in the basic science field as well as become an industry standard in applied drug discovery setting. With mass spec-based detection the CETSA® Explore format offers an unprecedented discovery tool for understanding the mode of action of small molecule drugs and biologics. In addition, thermal profiling of cells can help describe different cellular stress responses as it gives insight into the rewiring of protein-protein networks inside the cell upon stimuli. In the work presented here, we have used the Protein Integral Solubility alteration version of the CETSA® protocol (cCETSA®/PISA) to profile cellular responses to several hundred well described probes and marketed drugs.

TEA and the Cellular Context
In general, CETSA® fingerprints seem to correlate well with the known mechanism of action of compounds but also provide novel insight into less characterised compounds. To study how biological context affects CETSA® fingerprints, we’ve profiled a set of compounds in three different cell lines: HepG2 (hepatocellular carcinoma, epithelial-like), U87MG (glioblastoma, epithelial) and K562 (myeloid leukemia, lymphoblast).
How big is CETSA® fingerprints database? Download TEA poster.
References
- Martinez Molina, D. et al. Monitoring Drug Target Engagement in Cells and Tissues Using the Cellular Thermal Shift Assay. Science 341, 84-87 (2013)
- Savitski M. M., Reinhard F. B. M., Franken H., Werner T., Savitski M. F., Eberhard D., et al. (2014). Tracking Cancer Drugs in Living Cells by Thermal Profiling of the Proteome. Science 346 (6205), 1255784.
- Gaetani M., Sabatier P., Saei A. A., Beusch C. M., Yang Z., Lundström S. L., et al. (2019). Proteome Integral Solubility Alteration: A High-Throughput Proteomics Assay for Target Deconvolution. J. Proteome Res. 18 (11), 4027–4037.
- Friman at al., SLAS Discovery 2021, DOI: 10.1177/2472555220973597
- Chernobrovkin at al., SLAS Discovery 2021, DOI: 10.1177/2472555220984372
- Hendricks at al., ACS Chem. Biol. 2022, DOI: 10.1021/ acschembio.1c00488
